Code
# Load the iris dataset
data(iris)
# Perform Fisher's discriminant analysis
library(MASS)
lda_model <- lda(Species ~ ., data = iris)
# Print the summary of the analysis
print(lda_model)Call:
lda(Species ~ ., data = iris)
Prior probabilities of groups:
setosa versicolor virginica
0.3333333 0.3333333 0.3333333
Group means:
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.006 3.428 1.462 0.246
versicolor 5.936 2.770 4.260 1.326
virginica 6.588 2.974 5.552 2.026
Coefficients of linear discriminants:
LD1 LD2
Sepal.Length 0.8293776 0.02410215
Sepal.Width 1.5344731 2.16452123
Petal.Length -2.2012117 -0.93192121
Petal.Width -2.8104603 2.83918785
Proportion of trace:
LD1 LD2
0.9912 0.0088 Code
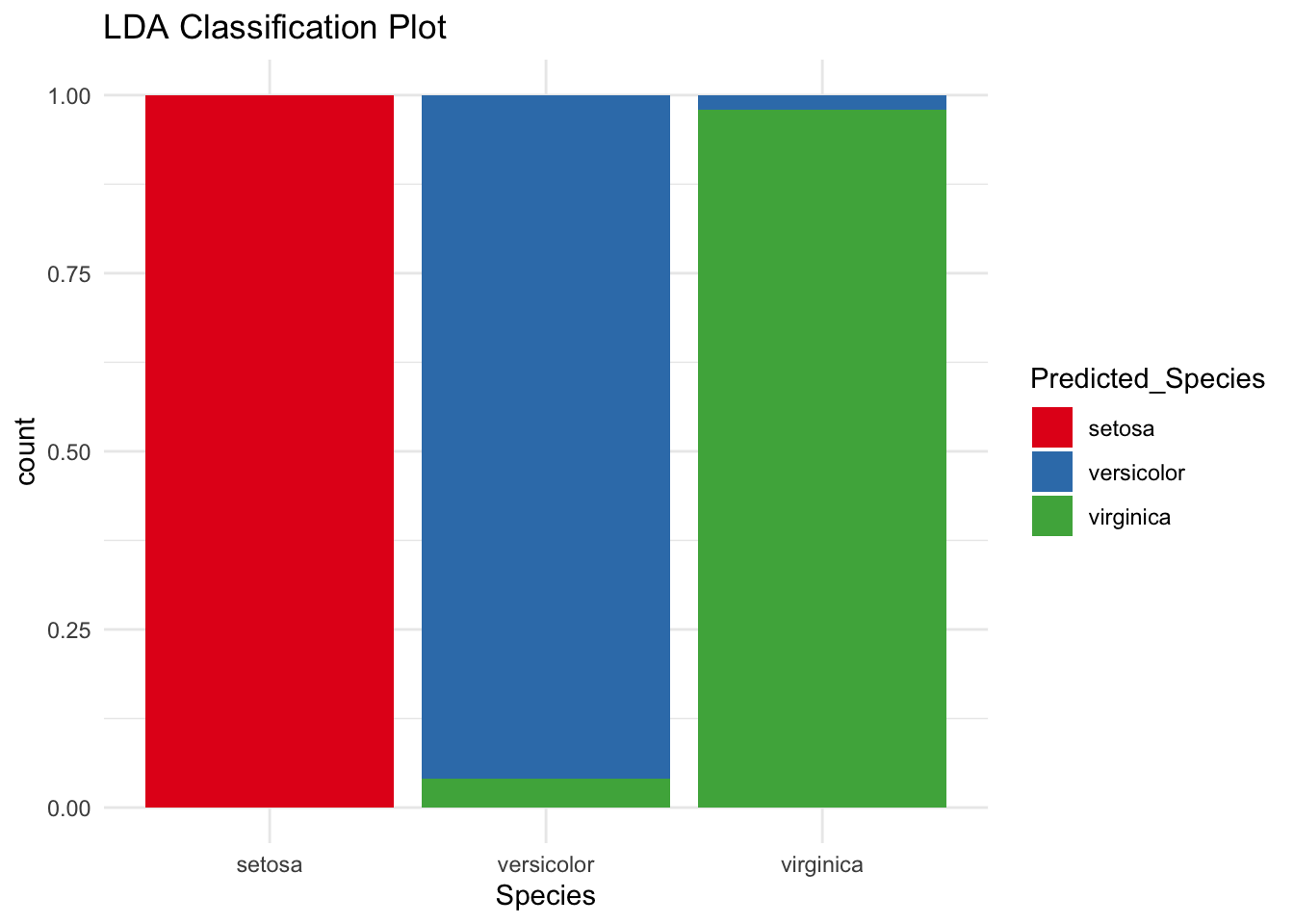
# Predict the species using the model
predicted_species <- predict(lda_model, iris)$class
# Compare the predicted species with the actual species
accuracy <- mean(predicted_species == iris$Species)
cat("Accuracy:", accuracy * 100, "%\n")Accuracy: 98 %