PsychENCODE Models
PsychENCODE analysis
Gandal et al analyzed autism spectrum disorder, schizophrenia, and bipolar disorder across multiple levels of transcriptomic organization—gene expression, local splicing, transcript isoform expression, and coexpression networks for both protein-coding and noncoding genes to produce a quantitative, genome-wide resource. They performed TWAS based on 2,188 postmortem frontal and temporal cerebral cortex samples from 1,695 adults. RNA-sequencing reads were aligned to the GRCh37.p13 (hg19) reference genome.
Gene expression level prediction models
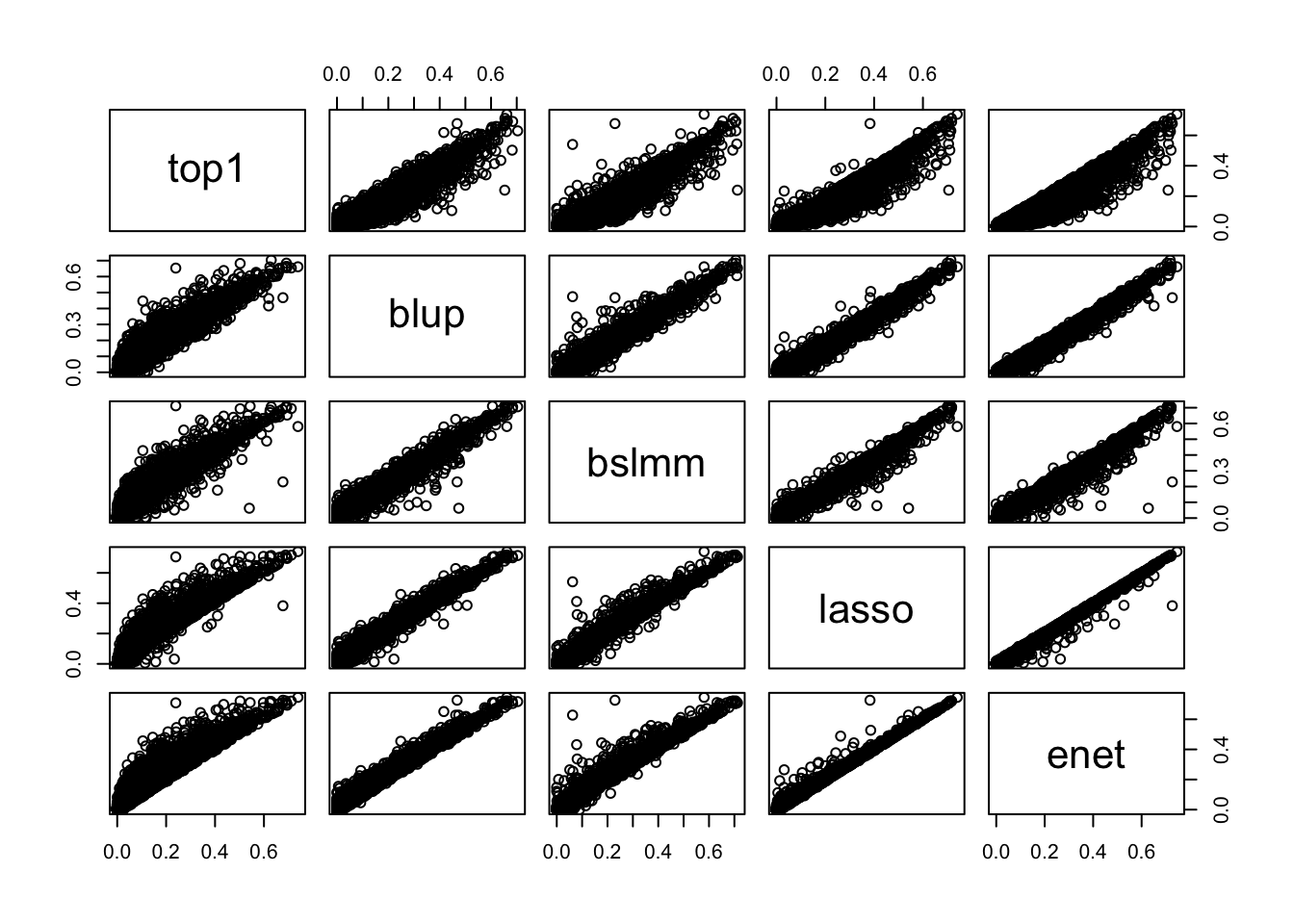
We extracted the elastic-net weights released provided Gandal et al. who chose (as prescribed by FUSION) the model that maximized the prediction performance among elastic net, BSLMM, lasso, top eQTL, and BLUP (best linear unbiased prediction, similar to ridge regression). Our extensive analysis had shown that this was not necessary since the very sparse architecture of gene expression traits were optimally predicted using elastic net in this paper. We further confirmed our assersion by comparing the different prediction model performance as provided by Gandal et al as shown in the figure below: the last column compares elastic net to all the other models and most points are either on or below the identity line.
More info on the study: https://science.sciencemag.org/content/362/6420/eaat8127. Their results were shared at http://resource.psychencode.org. Gandal et al’s transcriptome prediction models can be downloaded with this link PsychENCODE weights data link.
We used the weights they generated from the elastic net method and reformated it into a model compatible with PrediXcan software (code). We also calculated covariances between variants (code). The models can be downloaded here.
We validated the model by running PrediXcan on 1000G genotypes PsychENCODE, GTEx v7 Brain Cortex, and GTEx v7 Whole Blood tissue models, and then comparing the correlation between predicted gene expression and observed expression from GEUVADIS. The results can be found here.
Next, we compared PsychENCODE S-PrediXcan association results with GTEx v7 Brain Cortex and Whole Blood tissue models from the Walters Group Schizophrenia GWAS. The results can be found here.
The original PsychENCODE model was defined in hg19, but we also lifted it over to hg38. We validated it by comparing S-PrediXcan associations results from the original hg19, lifted over hg38, and GTEx v8 mashr Brain Cortex models on the Schizophrenia GWAS. The steps to lift over the model and the validation results are here.